
Nagy G, Oostenbrink C (2014) Dihedral-based segment identification and classification of biopolymers I: proteins. J Chem Inf Model 54(7):2166–2179įodje MN, Al-Karadaghi S (2002) Occurrence, conformational features and amino acid propensities for the π-helix.
Zacharias J, Knapp EW (2014) Protein secondary structure classification revisited: processing DSSP information with PSSC. Ramachandran GT, Sasisekharan V (1968) Conformation of polypeptides and proteins.

Proteins Struct Funct Bioinf 23(4):566–579 Biopolymers 22(12):2577–2637įrishman D, Argos P (1995) Knowledge-based protein secondary structure assignment. Kabsch W, Sander C (1983) Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Onuchic JN, Wolynes PG (2004) Theory of protein folding. John Wiley & Sons, Inc, Hoboken, New Jersey, pp 703–724Īnfinsen CB (1973) Principles that govern the folding of protein chains. BMC Bioinform 19(4):99–109Ībbass J, Nebel JC, Mansour N, Elloumi M, Zomaya AY (2013) Ab initio protein structure prediction: methods and challenges. Zhou J, Wang H, Zhao Z, Xu R, Lu Q (2018) CNNH_PSS: protein 8-class secondary structure prediction by convolutional neural network with highway. Proc Natl Acad Sci 96(25):14258–14263Įisenberg D (2003) The discovery of the α-helix and β-sheet, the principal structural features of proteins. Srinivasan R, Rose GD (1999) A physical basis for protein secondary structure. Encyclopedia of Bioin-formatics and Computational Biology, pp 488–496 Reeb J, Rost B (2019) Secondary structure prediction. Pauling L, Corey RB, Branson HR (1951) The structure of proteins: two hydrogen-bonded helical configurations of the polypeptide chain. Results from the fragment-based studies demonstrate the feasibility of applying deep learning solutions for structure assignment problems. The model has been successfully tested on a few full-length proteins also. The model uses only C α coordinates for secondary structure assignments. The model implemented in this work is trained with a subset of the protein fragments and achieves 88.1% and 82.5% train and test accuracy, respectively. A fast and efficient GPU-based parallel procedure extracts fragments from protein files.
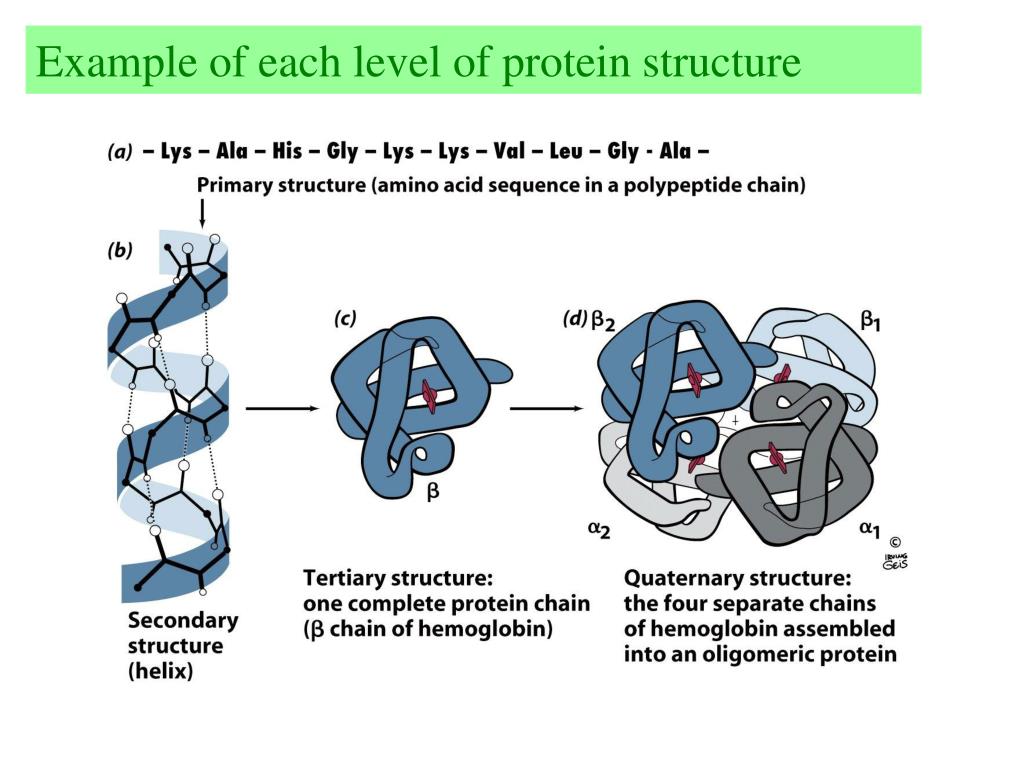
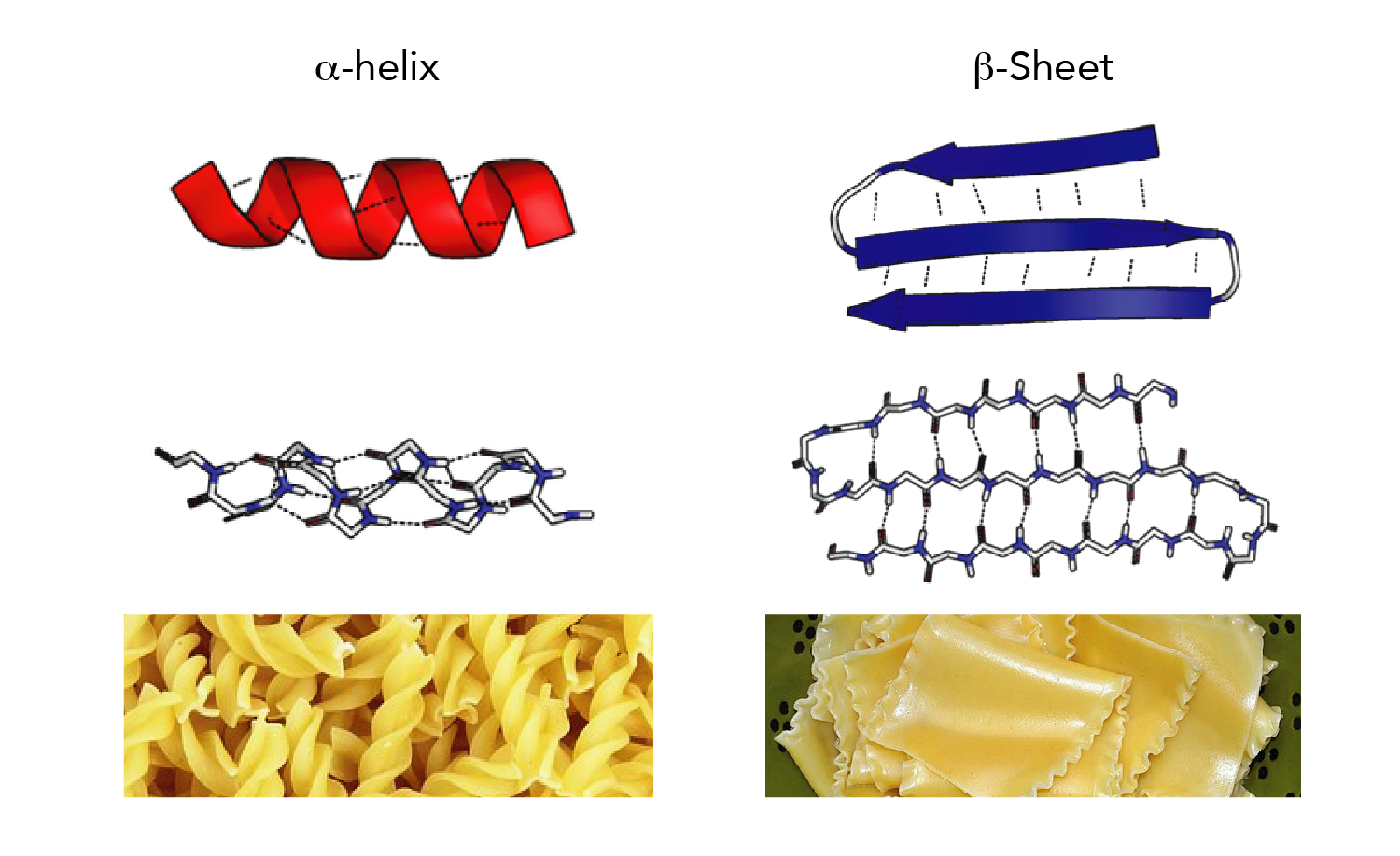
Our method proposed a multi-class classifier program named DLFSA for assigning protein secondary structure elements (SSE) using convolutional neural networks (CNNs). However, the assignment process becomes challenging when missing atoms are present in the protein files. Since the 1980s, various methods based on hydrogen bond analysis and atomic coordinate geometry, followed by machine learning, have been employed in protein structure assignment. Accurate and reliable structure assignment data is crucial for secondary structure prediction systems. Knowledge about protein structure assignment enriches the structural and functional understanding of proteins.


 0 kommentar(er)
0 kommentar(er)
